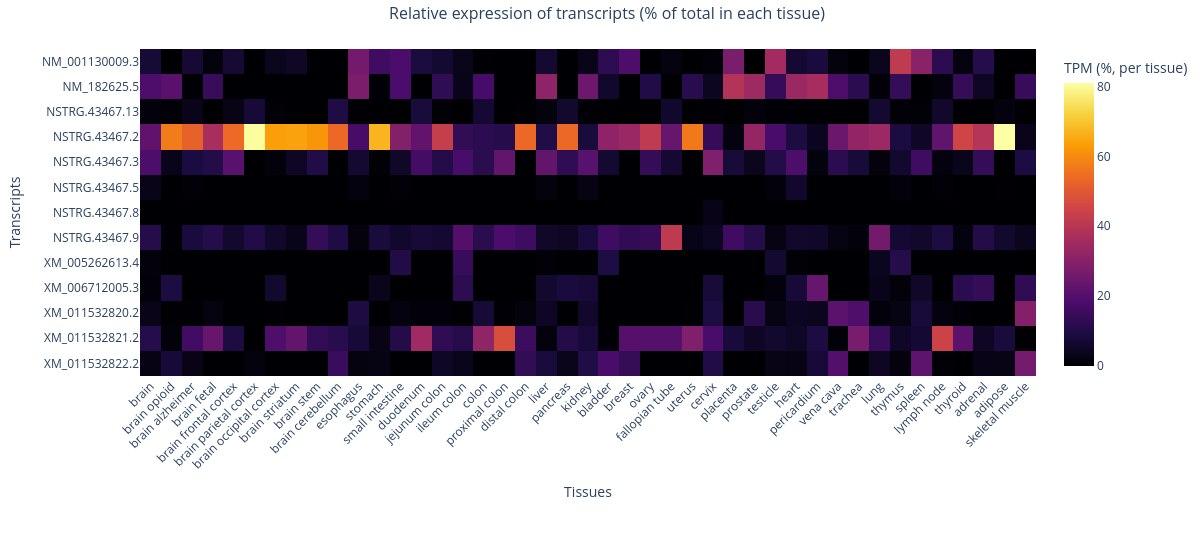
CRU 337 Phenotime Webtool
Database for queries of melanoma cohorts with molecular and clinical data
We provide a new database connecting whole transcriptomes with clinical data for straight-forward access and analysis of patient-specific samples. Users can perform association tests of survival and gene expression across different cohorts, identify cell-type expressions, or correlate the presence of immune cells.
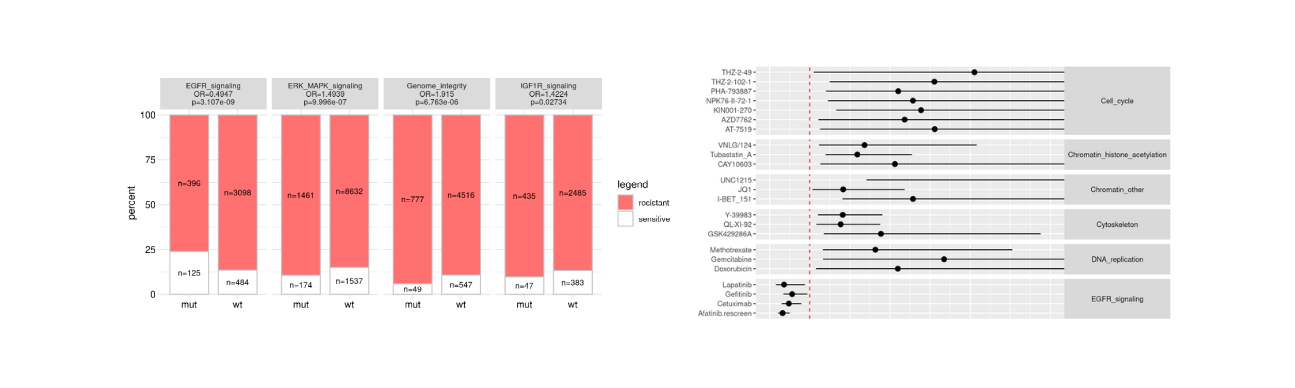
GDSC drug Response tool
Drug response test with pathways and forestplots for over 200 compounds
Test drug response for groups of cancer cell lines from the GDSC dataset. Test genetically changed vs. wild type. The grouping of cell lines for comparison will be based on genetic data at www.cBioportal.org.
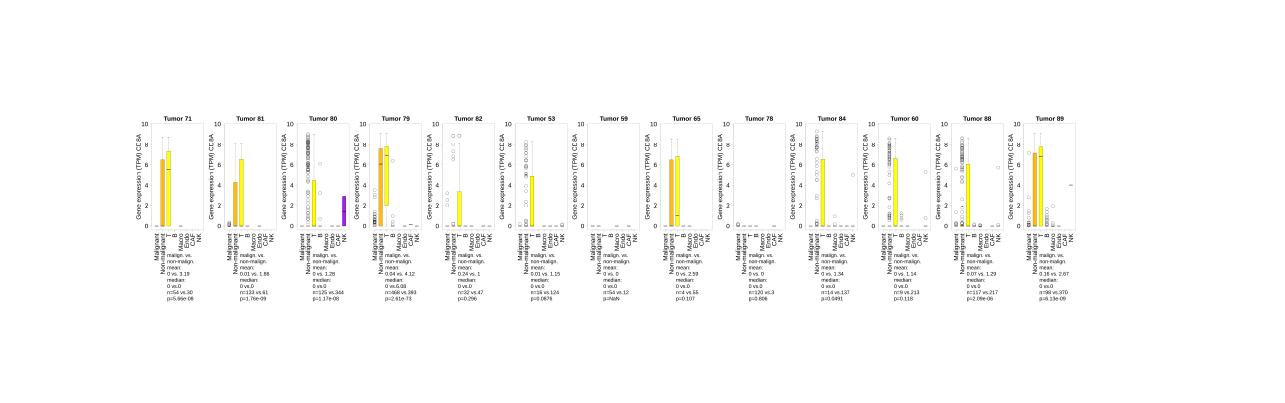
Single cell gene expression tool, melanoma
Query data from Tirosh et al. 2016
Compare gene expression in malignant and non-malignant cells of melanoma tumors.
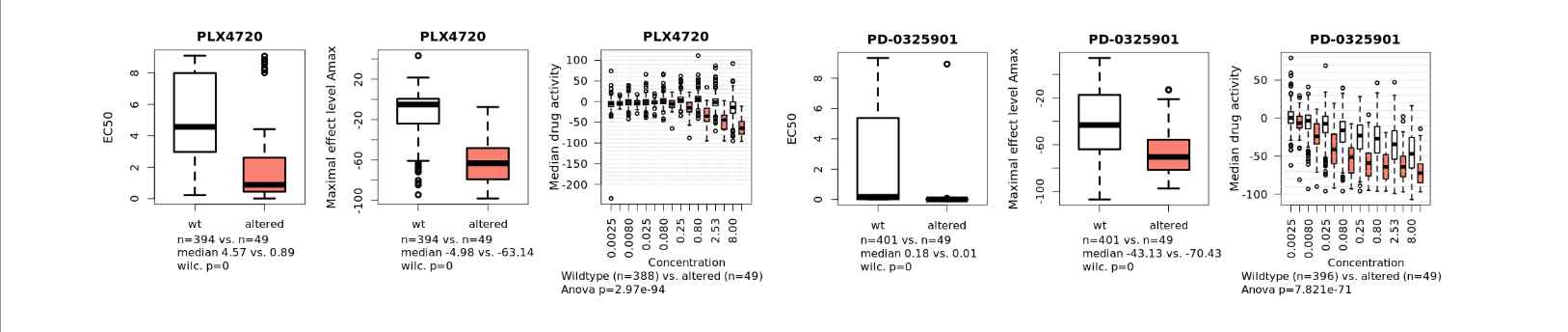
CCLE drug response tool
Drug response test with concentration response curves for over 20 compounds
Test drug response for groups of cancer cell lines from the CCLE dataset. Test genetically changed vs. wild type. The grouping of cell lines for comparison will be based on genetic data at www.cBioportal.org.